 ©2026 Biological and Chemical Oceanography Data Management Office.
©2026 Biological and Chemical Oceanography Data Management Office.Funded by the U.S. National Science Foundation
Marine sediments contain a microbial population large enough to rival that of Earth's oceans, but much about this vast community is unknown. Innovations in total cell counting methods have refined estimates of cell concentrations, but tell us nothing about specific taxa. Isotopic data provides evidence that a majority of subsurface microorganisms survive by breaking down organic matter, yet measurable links between specific microbial taxa and their organic matter substrates are untested. The proposed work overcomes these limitations, with a particular focus on the degradation of proteins and carbohydrates, which comprise the bulk of classifiable sedimentary organic matter. The project will link specific taxa to potential extracellular enzyme activity in the genomes of single microbial cells, apply newly-identified, optimal methods for counting viable cells belonging to specific taxa using catalyzed reporter deposition fluorescent in situ hybridization (CARD-FISH), and measure the potential activity of their enzymes in situ. The resulting data will provide key evidence about the strategies subsurface life uses to overcome extreme energy limitation and contribute to the long-term carbon cycle.
The Principal Investigators are employing novel,improved methods to quantify cells of specific taxa in the marine subsurface and to determine the biogeochemical functions of those uncultured taxa, including:
1) Determine the pathway of organic carbon degradation in single cell genomes of uncultured, numerically dominant subsurface microorganisms.
2) Quantify viable bacteria and archaea in the deep subsurface using an improvement on the existing technology of CARD-FISH.
3 )Measure the potential activities (Vmax values) of enzymes in deep Baltic Sea sediments, and use the abundances of enzyme-producing microorganisms to calculate depth profiles of cell-specific Vmax values.
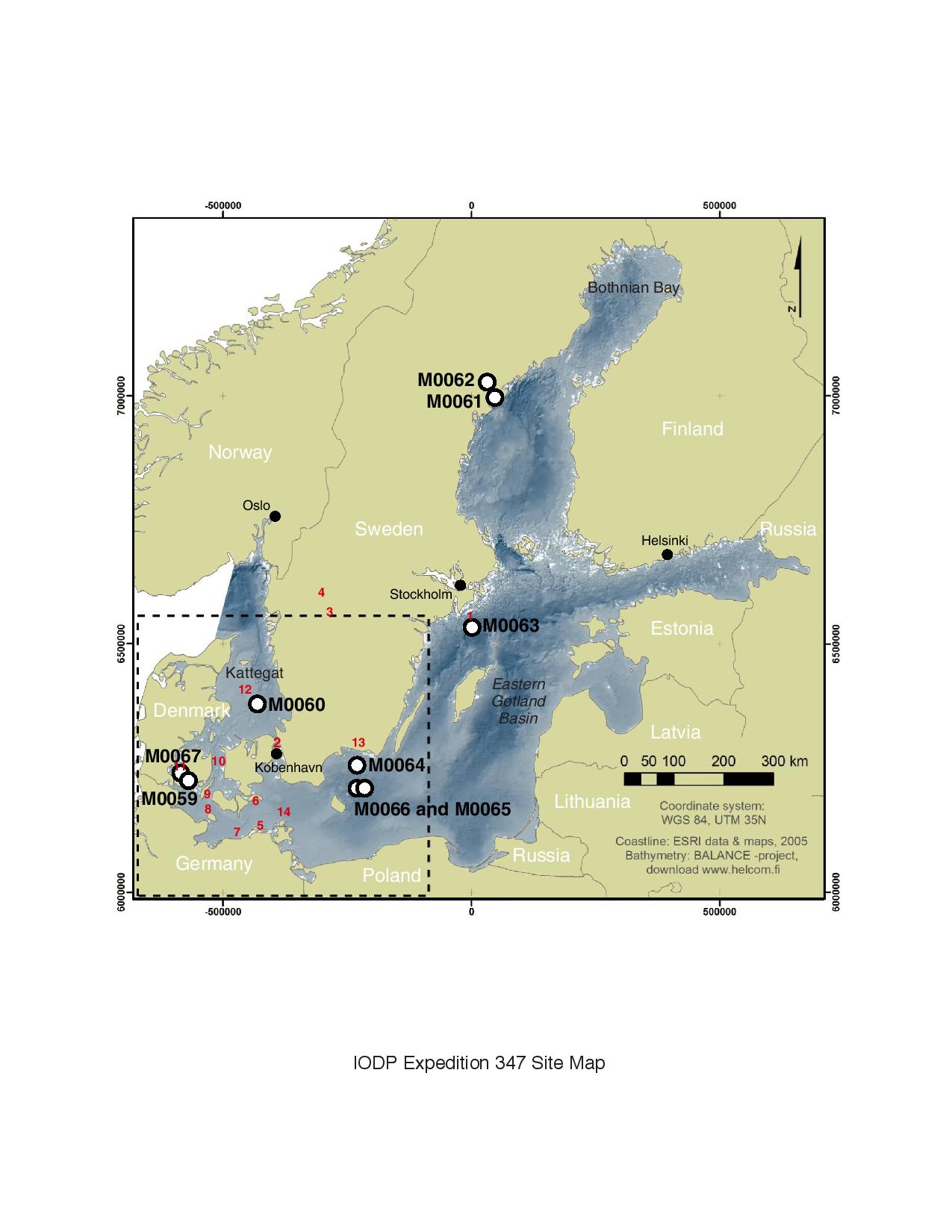
The project combines these methods in order to identify and quantify the cells capable of degrading organic matter in deep sediments of the Baltic Sea, obtained from Integrated Ocean Drilling Program (IODP) expedition 347. These results will greatly expand our knowledge of the function and activity of uncultured microorganisms in the deep subsurface.
This project is associated with C-DEBI account number 157595.
Additional data for this site are managed by and directly available from the project data site: http://iodp.pangaea.de/front_content.php?idcat=556
| Dataset | Latest Version Date | Current State |
|---|---|---|
| Microbial diversity and geochemistry of marine sediment mesocosm, Cape Lookout Bight, North Carolina | 2016-07-27 | Final no updates expected |
| Methane and sulfate concentration profiles - sediment cores from White Oak River estuary in October 2012 (IODP-347 Microbial Quantification project) | 2016-06-22 | Final no updates expected |
| Quantitative PCR data from sediment samples from MPSV GREATSHIP MANISHA IODP-347 cruise in the Baltic Sea in 2013 (IODP-347 Microbial Quantification project) | 2016-03-25 | Final no updates expected |
| Drill site locations from MPSV GREATSHIP MANISHA IODP-347 cruise in the Baltic Sea in 2013 (IODP-347 Microbial Quantification project) | 2016-03-24 | Final no updates expected |
Principal Investigator: Karen G. Lloyd
University of Tennessee Knoxville (UTK)
Co-Principal Investigator: Andrew Steen
University of Tennessee Knoxville (UTK)
Contact: Karen G. Lloyd
University of Tennessee Knoxville (UTK)
Center for Dark Energy Biosphere Investigations [C-DEBI]
International Ocean Discovery Program [IODP]
DMP_20140122_KarenGLloyd.pdf (98.83 KB)
02/29/2016